Data pipelines
In essence, an ETL (Extract, Transform, Load) process is a Directed Acyclic Graph (DAG) of jobs where each job reads data, performs computations, and outputs new or updated datasets.
Windmill streamlines the creation of data pipelines that are not only fast and reliable but also straightforward to construct:
- Developer Experience: Windmill's design facilitates the swift assembly of data flows, allowing for step-by-step data processing in a visually intuitive and manageable manner.
- Control and Efficiency: It offers the ability to manage parallelism across steps and set concurrency limits to accommodate external resources that may be sensitive to overload or have rate limits.
- Flexibility in Execution: Flows in Windmill can be restarted from any point, enhancing the process of pipeline development and debugging by making it more flexible and efficient.
- Simplified Monitoring: Built-in error and recovery handling mechanisms simplify monitoring, ensuring that managing your data pipelines is both effective and straightforward.
The particularity of data pipeline flows vs. any other kind of automation flows is that they run computation on large datasets and the result of such computation is itself a (potentially large) dataset that needs to be stored.
For the compute, as data practitioner for the most demanding ETLs, we have observed that in almost all cases, the system they run on is ill-designed for their task. Much faster alternatives now exist leveraging the modern OLAP processing libraries. We have integrated with Polars and DuckDB, as ones of the best-in-class in-memory data processing libraries and they fit particularly well Windmill since you can assign variously sized workers depending on the step.
To give you a quick idea:
- Running a
SELECT COUNT(*), SUM(column_1), AVG(column_2) FROM my_table GROUP_BY keywith 600M entries inmy_tablerequires less than 24Gb of memory using DuckDB - Running a
SELECT * FROM table_a JOIN table_b ORDER BY key, withtable_ahaving 300M rows andtable_b75M rows with DuckDB requires 24Gb of memory
Add to those numbers that on AWS for example, you can get up to 24Tb of memory on a single server. Nowadays, you don't need a complex distributed computing architecture to process a large amount of data.
And for storage, you can now link a Windmill workspace to an S3 bucket and use it as source and/or target of your processing steps seamlessly, without any boilerplate.
The very large majority of ETLs can be processed step-wise on single nodes and Windmill provides (one of) the best models for orchestrating non-sharded compute. Using this model, your ETLs will see a massive performance improvement, your infrastructure will be easier to manage and your pipeline will be easier to write, maintain, and monitor.
Windmill integration with an external object storage
In Windmill, a data pipeline is implemented using a flow, and each step of the pipeline is a script. One of the key features of Windmill flows is to easily pass a step result to its dependent steps. But because those results are serialized to Windmill database and kept as long as the job is stored, this obviously won't work when the result is a dataset of millions of rows. The solution is to save the datasets to an external storage at the end of each script.
In most cases, S3 is a well-suited storage and Windmill now provides a basic yet very useful integration with external S3 storage at the workspace level.
The first step is to define an S3 resource in Windmill and assign it to be the Workspace S3 bucket in the workspace settings.
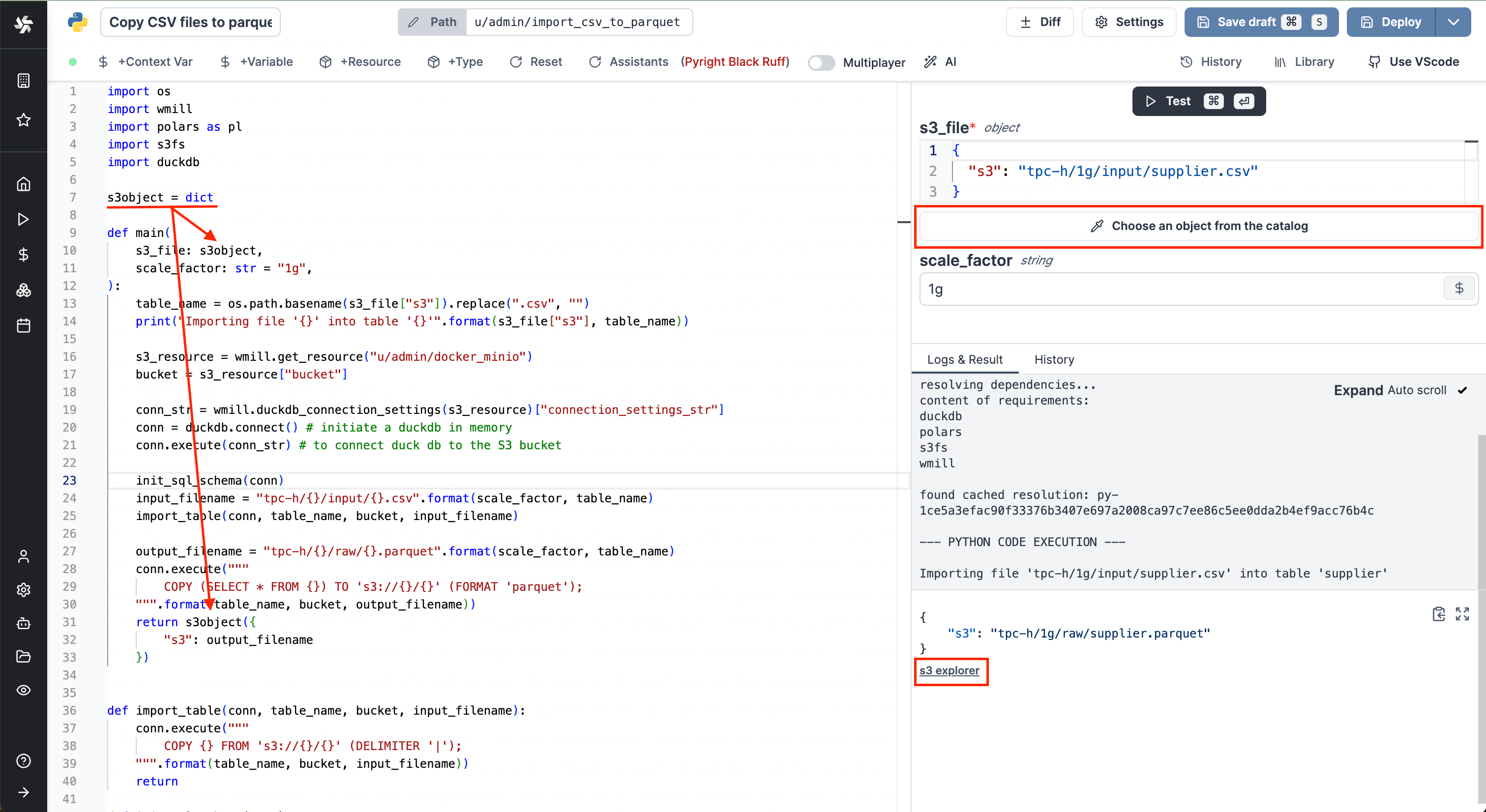
From now on, Windmill will be connected to this bucket and you'll have easy access to it from the code editor and the job run details. If a script takes as input a s3object, you will see in the input form on the right a button helping you choose the file directly from the bucket.
Same for the result of the script. If you return an s3object containing a key s3 pointing to a file inside your bucket, in the result panel there will be a button to open the bucket explorer to visualize the file.
S3 files in Windmill are just pointers to the S3 object using its key. As such, they are represented by a simple JSON:
{
"s3": "path/to/file"
}
Clicking on the button will lead directly to a bucket explorer. You can browse the bucket content and even visualize file content without leaving Windmill.
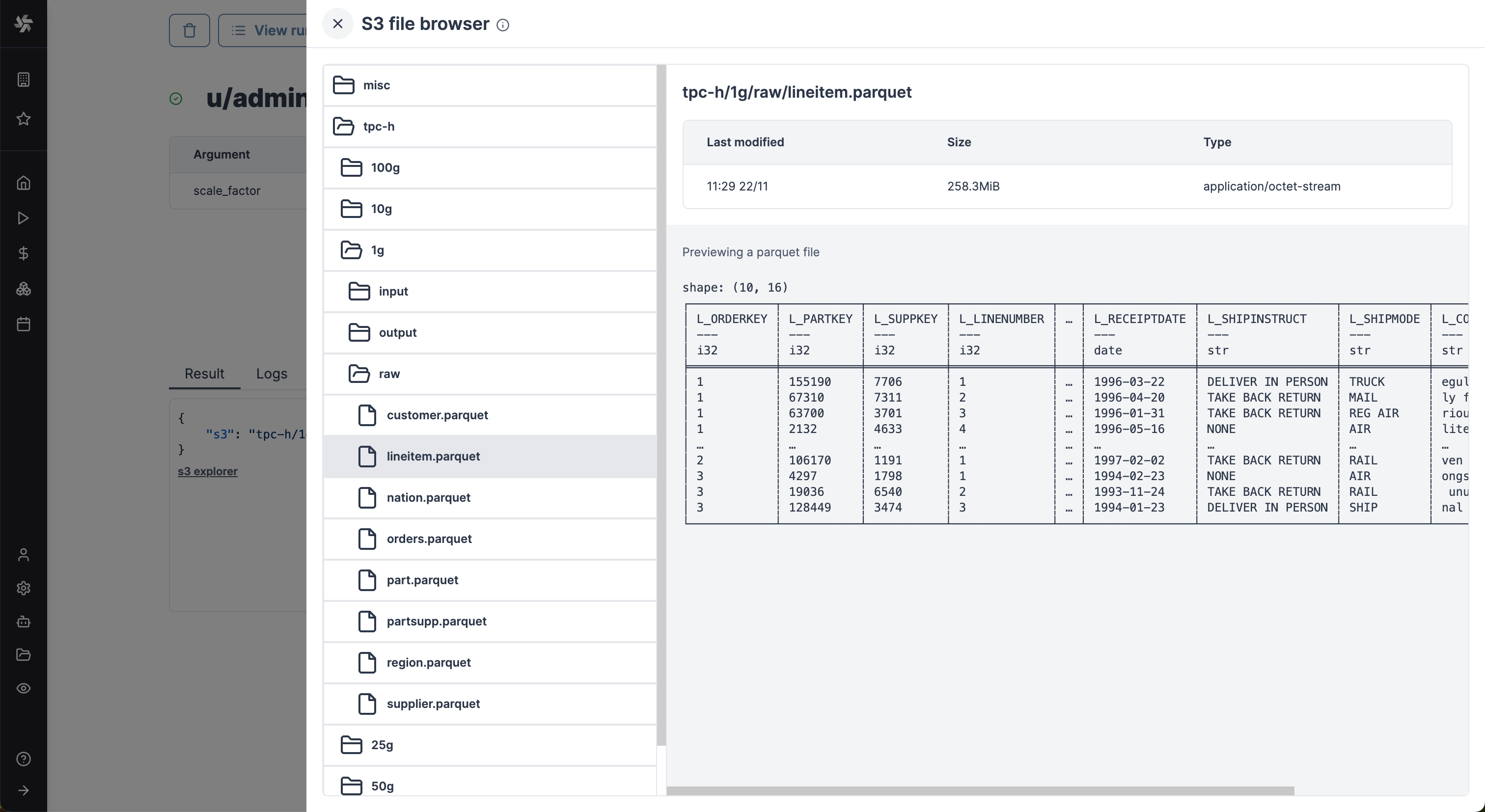
Clicking on one of those buttons, a drawer will open displaying the content of the workspace bucket. You can select any file to get its metadata and if the format is common, you'll see a preview. In the above picture, for example, we're showing a Parquet file, which is very convenient to quickly validate the result of a script.
From there you always have the possibility to use the S3 client library of your choice to read and write to S3. That being said, Polars and DuckDB can read/write directly from/to files stored in S3 Windmill now ships with helpers to make the entire data processing mechanics very cohesive.
Find all details at:
Windmill integration with Polars and DuckDB for data pipelines
ETLs can be easily implemented in Windmill using its integration with Polars and DuckDB for facilitate working with tabular data. In this case, you don't need to manually interact with the S3 bucket, Polars/DuckDB does it natively and in a efficient way. Reading and Writing datasets to S3 can be done seamlessly.
- Polars
- DuckDB
#requirements:
#polars==0.20.2
#s3fs==2023.12.0
#wmill>=1.229.0
import wmill
from wmill import S3Object
import polars as pl
import s3fs
def main(input_file: S3Object):
bucket = wmill.get_resource("<PATH_TO_S3_RESOURCE>")["bucket"]
# this will default to the workspace S3 resource
storage_options = wmill.polars_connection_settings().storage_options
# this will use the designated resource
# storage_options = wmill.polars_connection_settings("<PATH_TO_S3_RESOURCE>").storage_options
# input is a parquet file, we use read_parquet in lazy mode.
# Polars can read various file types, see
# https://pola-rs.github.io/polars/py-polars/html/reference/io.html
input_uri = "s3://{}/{}".format(bucket, input_file["s3"])
input_df = pl.read_parquet(input_uri, storage_options=storage_options).lazy()
# process the Polars dataframe. See Polars docs:
# for dataframe: https://pola-rs.github.io/polars/py-polars/html/reference/dataframe/index.html
# for lazy dataframe: https://pola-rs.github.io/polars/py-polars/html/reference/lazyframe/index.html
output_df = input_df.collect()
print(output_df)
# To write back the result to S3, Polars needs an s3fs connection
s3 = s3fs.S3FileSystem(**wmill.polars_connection_settings().s3fs_args)
output_file = "output/result.parquet"
output_uri = "s3://{}/{}".format(bucket, output_file)
with s3.open(output_uri, mode="wb") as output_s3:
# persist the output dataframe back to S3 and return it
output_df.write_parquet(output_s3)
return S3Object(s3=output_file)
#requirements:
#wmill>=1.229.0
#duckdb==0.9.1
import wmill
from wmill import S3Object
import duckdb
def main(input_file: S3Object):
bucket = wmill.get_resource("u/admin/windmill-cloud-demo")["bucket"]
# create a DuckDB database in memory
# see https://duckdb.org/docs/api/python/dbapi
conn = duckdb.connect()
# this will default to the workspace S3 resource
args = wmill.duckdb_connection_settings().connection_settings_str
# this will use the designated resource
# args = wmill.duckdb_connection_settings("<PATH_TO_S3_RESOURCE>").connection_settings_str
# connect duck db to the S3 bucket - this will default to the workspace S3 resource
conn.execute(args)
input_uri = "s3://{}/{}".format(bucket, input_file["s3"])
output_file = "output/result.parquet"
output_uri = "s3://{}/{}".format(bucket, output_file)
# Run queries directly on the parquet file
query_result = conn.sql(
"""
SELECT * FROM read_parquet('{}')
""".format(
input_uri
)
)
query_result.show()
# Write the result of a query to a different parquet file on S3
conn.execute(
"""
COPY (
SELECT COUNT(*) FROM read_parquet('{input_uri}')
) TO '{output_uri}' (FORMAT 'parquet');
""".format(
input_uri=input_uri, output_uri=output_uri
)
)
conn.close()
return S3Object(s3=output_file)
Polars and DuckDB need to be configured to access S3 within the Windmill script. The job will need to accessed the S3 resources, which either needs to be accessible to the user running the job, or the S3 resource needs to be set as public in the workspace settings.
Canonical data pipeline in Windmill w/ Polars and DuckDB
With S3 as the external store, a transformation script in a flow will typically perform:
- Pulling data from S3.
- Running some computation on the data.
- Storing the result back to S3 for the next scripts to be run.
Windmill SDKs now expose helpers to simplify code and help you connect Polars or DuckDB to the Windmill workspace S3 bucket. In your usual IDE, you would need to write for each script:
conn = duckdb.connect()
conn.execute(
"""
SET home_directory='./';
INSTALL 'httpfs';
LOAD 'httpfs';
SET s3_url_style='path';
SET s3_region='us-east-1';
SET s3_endpoint='http://minio:9000'; # using MinIo in Docker works perfectly fine if you don't have access to an AWS S3 bucket!
SET s3_use_ssl=0;
SET s3_access_key_id='<ACCESS_KEY>';
SET s3_secret_access_key='<SECRET_KEY>';
"""
)
# then you can start using your connection to pull CSVs/Parquet/JSON/... files from S3
conn.sql("SELECT * FROM read_parquet(s3://windmill_bucket/file.parquet)")
In Windmill, you can just do:
conn = duckdb.connect()
s3_resource = wmill.get_resource("/path/to/resource")
conn.execute(wmill.duckdb_connection_settings(s3_resource)["connection_settings_str"])
conn.sql("SELECT * FROM read_parquet(s3://windmill_bucket/file.parquet)")
And similarly for Polars:
args = {
"anon": False,
"endpoint_url": "http://minio:9000",
"key": "<ACCESS_KEY>",
"secret": "<SECRET_KEY>",
"use_ssl": False,
"cache_regions": False,
"client_kwargs": {
"region_name": "us-east-1",
},
}
s3 = s3fs.S3FileSystem(**args)
with s3.open("s3://windmill_bucket/file.parquet", mode="rb") as f:
dataframe = pl.read_parquet(f)
becomes in Windmill:
s3_resource = wmill.get_resource("/path/to/resource")
s3 = s3fs.S3FileSystem(**wmill.polars_connection_settings(s3_resource))
with s3.open("s3://windmill_bucket/file.parquet", mode="rb") as f:
dataframe = pl.read_parquet(f)
And more to come! With both Windmill providing the boilerplate code, and Polars and DuckDB handling reading and writing from/to S3 natively, you can interact with S3 files very naturally and your Windmill scripts become concise and focused on what really matters: processing the data.
In the end, a canonical pipeline step in Windmill will look something like this:
import polars as pl
import s3fs
import datetime
import wmill
s3object = dict
def main(input_dataset: s3object):
# initialization: connect Polars to the workspace bucket
s3_resource = wmill.get_resource("/path/to/resource")
s3 = s3fs.S3FileSystem(wmill.duckdb_connection_settings(s3_resource))
# reading data from s3:
bucket = s3_resource["bucket"]
input_dataset_uri = "s3://{}/{}".format(bucket, input_dataset["s3"])
output_dataset_uri = "s3://{}/output.parquet".format(bucket)
with s3.open(input_dataset_uri, mode="rb") as input_dataset, s3.open(output_dataset_uri, mode="rb") as output_dataset:
input = pl.read_parquet(input_dataset)
# transforming the data
output = (
input.filter(pl.col("L_SHIPDATE") >= datetime.datetime(1994, 1, 1))
.filter(
pl.col("L_SHIPDATE")
< datetime.datetime(1994, 1, 1) + datetime.timedelta(days=365)
)
.filter((pl.col("L_DISCOUNT").is_between(0.06 - 0.01, 0.06 + 0.01)))
.filter(pl.col("L_QUANTITY") < 24)
.select([(pl.col("L_EXTENDEDPRICE") * pl.col("L_DISCOUNT")).alias("REVENUE")])
.sum()
.collect()
)
# writing the output back to S3
output.write_parquet(output_dataset)
# returning the URI of the output for next steps to process it
return s3object({
"s3": output_dataset_uri
})
The example uses Polars. If you're more into SQL you can use DuckDB, but the code will have the same structure: initialization, reading from S3, transforming, writing back to S3.
In-memory data processing performance
By using Polars, DuckDB, or any other data processing libraries inside Windmill, the computation will happen on a single node. Even though you might have multiple Windmill workers, a script will still be run by a single worker and the computation won't be distributed. We've run some benchmarks to expose the performance and scale you could expect for such a setup.
We've taken a well-known benchmark dataset: the TCP-H dataset. It has the advantage of being available in any size and being fairly representative of real use cases. We've generated multiple versions: 1Gb, 5Gb, 10Gb and 25Gb (if you prefer thinking in terms of rows, the biggest table of the 25Gb version has around 150M rows). We won't detail here the structure of the database or the queries we've run, but TPC-H is well-documented if needed.
The following procedure was followed:
- Datasets provided by TPC-H as CSVs were uploaded as parquet files on S3.
- TPC-H provides a set of canonical queries. They perform numerous joins, aggregations, group-bys, etc. 8 of them were converted in the different dialects.
- Those queries were run sequentially as scripts in for-loop flow in Windmill, and this for each of the benchmark sets of data (1Gb, 5Gb, 10Gb, etc.). The memory of the Windmill server was recorded.
- Each script was:
- Reading the data straight from the S3 parquet files
- Running the query
- Storing the result in a separate parquet file on S3
A couple of notes before the results:
- We've run those benchmarks on a
m4.xlargeAWS server (8 vCPUs, 32Gb of memory). It's not a small server, but also not terribly large. Keep in mind you can get up to 24Tb of Memory on a single server on AWS (yes, it's not cheap, but it's possible!) - Polars comes with a lazy mode, in which it is supposed to be more memory efficient. We've benchmarked both normal and lazy mode.
- We also ran those benchmarks on Spark, as a well-known and broadly used reference. To be as fair as possible, the Spark "cluster" was composed of a single node running also on an
m4.xlargeAWS instance.
Polars is the fastest at computing the results, but consumes slightly more memory than Spark (it OOMed for the 25G benchmark). Polars in lazy mode, however, is a lot more memory efficient and can process more data, at the expense of computation time. Overall, both Polars and DuckDB behave very well in terms of memory consumption and computation time. The 10G benchmark contains tables with up to 60 million rows, and we were far from using the most powerful AWS instance. So, it is true that this doesn't scale horizontally, but it also confirms that a majority of data pipelines can be addressed with a large enough instance. And when you think about it, what's more convenient? Managing a single beefy server or a fleet of small servers?
DuckDB offers the possibility to back its database with a file on disk to save some memory. This mode fits perfectly with Windmill flows using a shared directory between steps. We implemented a simple flow where the first step loads the DB in a file, and the following steps consume this file to run the queries. We were able to run the 8 queries on the 100Gb benchmark successfully. It took 40 minutes and consumed 29,1Gb at the peak.
Limit of the number of executions per second
Windmill's core is its queue of jobs which is implemented in Postgres using the UPDATE SKIP LOCKED pattern. It can scale comfortably to 5k requests per second (RPS) on a normal Postgres database during benchmarks.